Research Interests
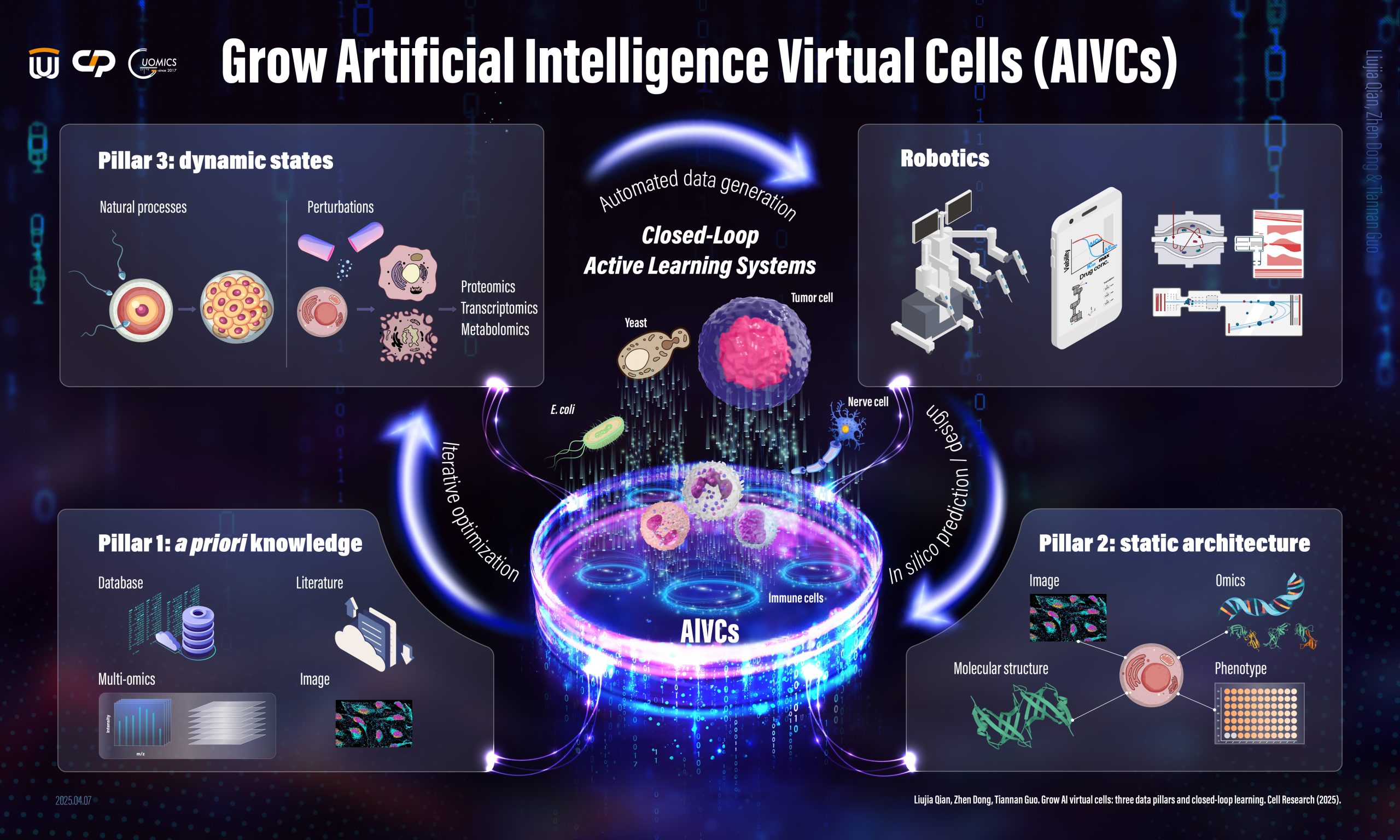
We are dedicated to constructing AI Virtual Cells to model and simulate cellular processes by leveraging spatial proteomics, perturbation proteomics, and other advanced technologies. Achieving this ambitious goal involves identifying the proteins expressed in cells and tissues, mapping their spatial distribution within subcellular organelles, analyzing proteome dynamics in response to perturbations, and integrating these insights through advanced AI modeling. Through this comprehensive approach, we strive to develop accurate and predictive virtual representations of cellular systems, ultimately advancing drug discovery and enhancing disease diagnosis.
We are also exploring the development of an AI-empowered mass spectrometer system.