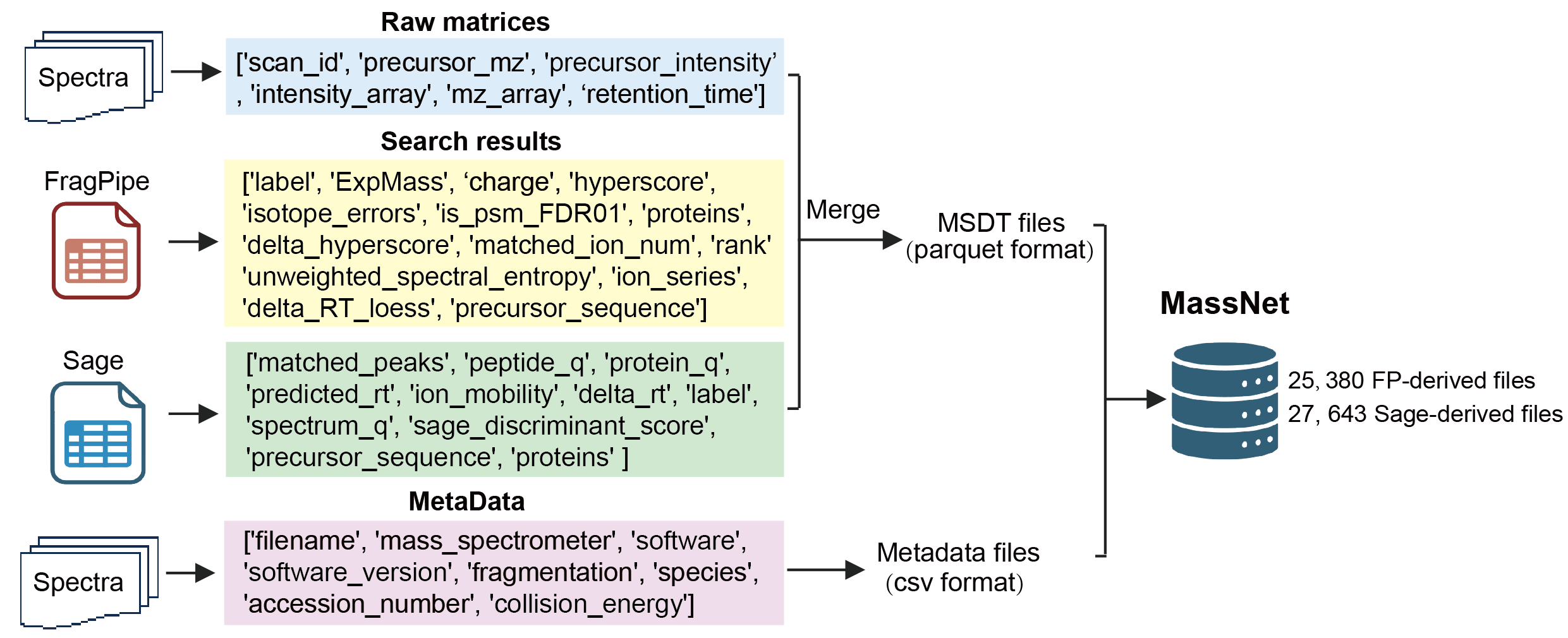
Mass Spectrometry Data Tensor (MSDT)
MassNet adopts a unified data representation strategy by structuring raw spectra into two-dimensional tensors that preserve key features such as mass-to-charge ratio (m/z) and intensity. All data are stored in Apache Parquet format, which offers advantages such as columnar storage, compression, and schema definition. This format enables efficient batch reading and integrates seamlessly with the DataLoader modules of mainstream deep learning frameworks, including PyTorch and TensorFlow, thereby streamlining data preprocessing. The ready-to-use format not only significantly accelerates model training on hardware accelerators such as GPUs and TPUs but also improves training reproducibility. By lowering the technical barrier for AI researchers and developers working with MS data, it substantially enhances the usability and scalability of deep learning methods in proteomics analysis. In addition, each spectrum is paired with comprehensive metadata, including instrument type, fragmentation method, sample origin, and corresponding PSM information, facilitating multi-dimensional and multi-scale modeling and analysis.